Produce summary tableGrobs as R graphics. For this functionality the suggested package gridExtra is required. To visualize the resulting object:
Usage
top_abund_tableGrob(
df,
id_cols = mandatory_IS_vars(),
quant_col = "fragmentEstimate_sum_PercAbundance",
by = "TimePoint",
alluvial_plot = NULL,
top_n = 10,
tbl_cols = "GeneName",
include_id_cols = FALSE,
digits = 2,
perc_symbol = TRUE,
transform_by = NULL
)Arguments
- df
A data frame
- id_cols
Character vector of id column names. To plot after alluvial, these columns must be the same as the
alluviaargument of integration_alluvial_plot.- quant_col
Column name holding the quantification value. To plot after alluvial, these columns must be the same as the
plot_yargument of integration_alluvial_plot.- by
The column name to subdivide tables for. The function will produce one table for each distinct value in
by. To plot after alluvial, these columns must be the same as theplot_xargument of integration_alluvial_plot.- alluvial_plot
Either NULL or an alluvial plot for color mapping between values of y.
- top_n
Integer. How many rows should the table contain at most?
- tbl_cols
Table columns to show in the final output besides
quant_col.- include_id_cols
Logical. Include
id_colsin the output?- digits
Integer. Digits to show for the quantification column
- perc_symbol
Logical. Show percentage symbol in the quantification column?
- transform_by
Either a function or a purrr-style lambda. This function is applied to the column
bybefore separating columns. IfNULLno function is applied. Useful to modify column order in final table.
Examples
data("integration_matrices", package = "ISAnalytics")
data("association_file", package = "ISAnalytics")
aggreg <- aggregate_values_by_key(
x = integration_matrices,
association_file = association_file,
value_cols = c("seqCount", "fragmentEstimate")
)
abund <- compute_abundance(x = aggreg)
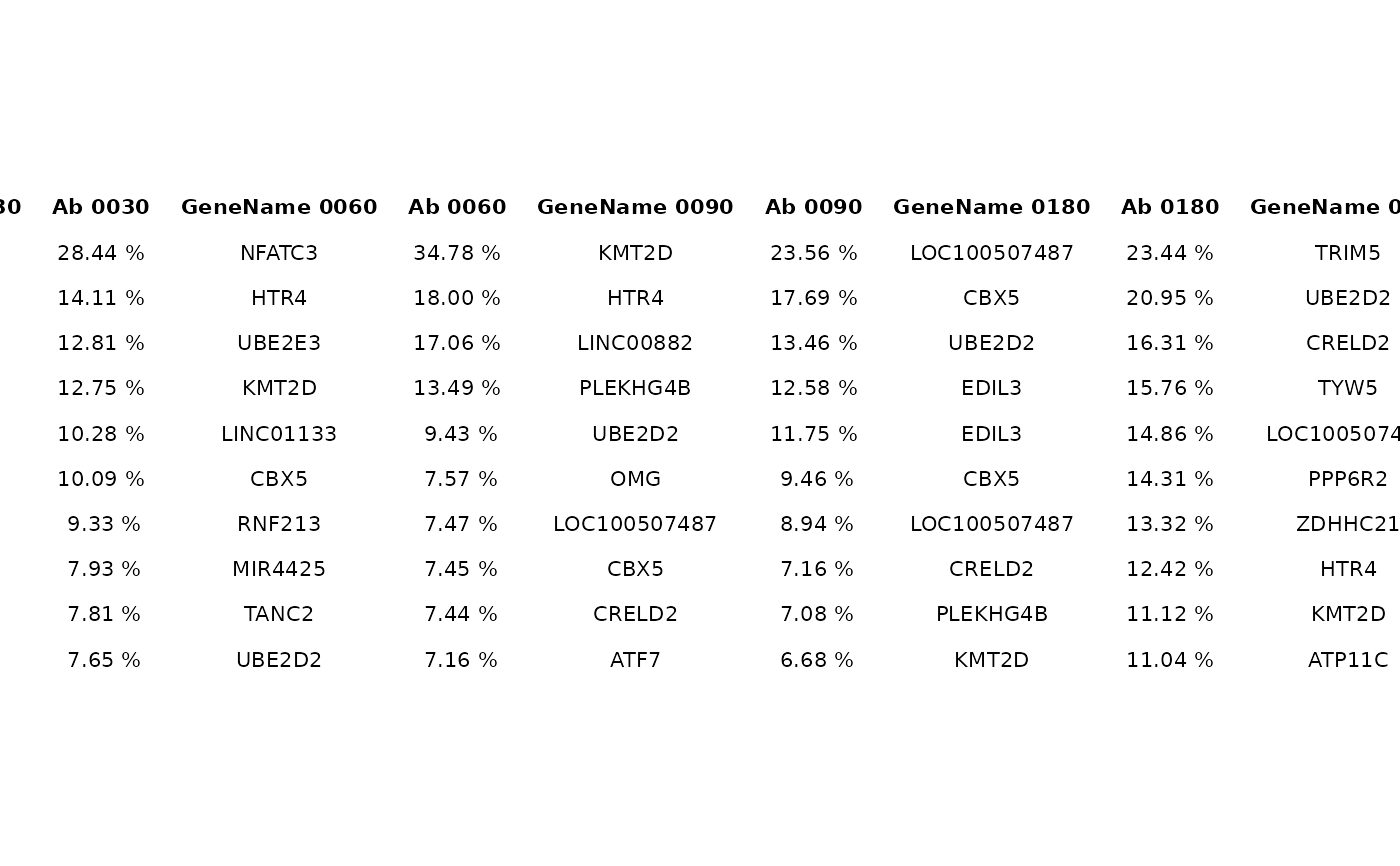
grob <- top_abund_tableGrob(abund)
gridExtra::grid.arrange(grob)
 # with transform
grob <- top_abund_tableGrob(abund, transform_by = ~ as.numeric(.x))
# with transform
grob <- top_abund_tableGrob(abund, transform_by = ~ as.numeric(.x))
